Lower Duncan River Kokanee AUC Analysis 2013
The suggested citation for this analytic report is:
Thorley, J.L. and Hogan, P.M. (2015) Lower Duncan River Kokanee AUC Analysis 2013. A Poisson Consulting Analysis Report. URL: https://www.poissonconsulting.ca/f/577555490.
Background
Since 2008 aerial surveys have been conducted in the Lower Duncan River (LDR) to count the number of kokanee spawners. The primary objectives of the current analysis report are to
- Estimate the aerial observer efficiency based on ground counts
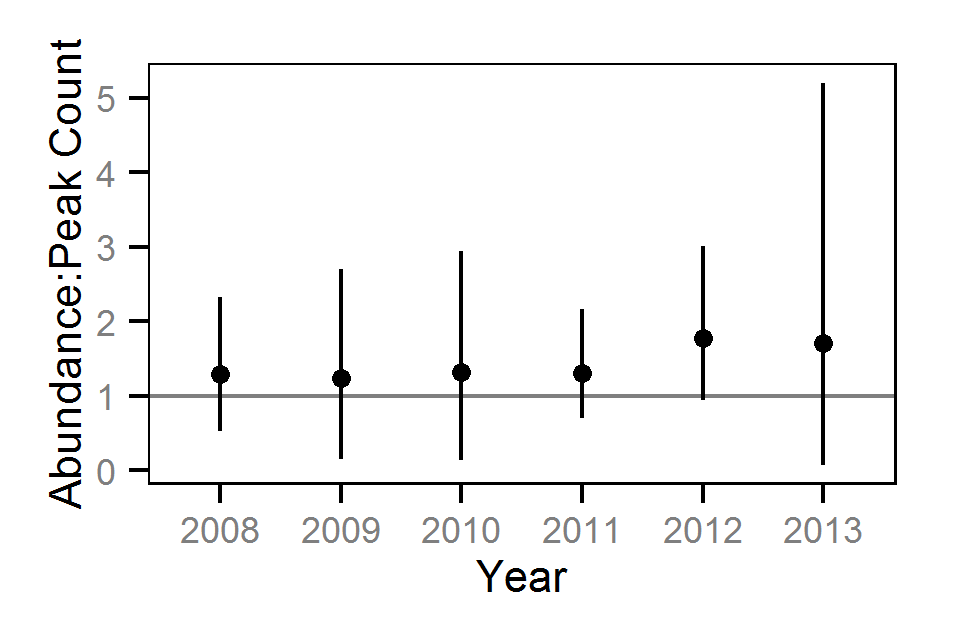
- Estimate the annual kokanee spawner abundance from the aerial counts
- Estimate the peak spawn timing
Methods
Data
The data were provided by LGL Limited in the form of an Access database and an Excel spreadsheet.
Analysis
Hierarchical Bayesian models were fitted to the LDR kokanee enumeration data using R version 3.0.2 (Team, 2013) and JAGS 3.3.0 (Plummer, 2012) which interfaced with each other via jaggernaut 1.6 (Thorley, 2014). For additional information on hierarchical Bayesian modelling in the BUGS language, of which JAGS uses a dialect, the reader is referred to Kery and Schaub (2011) pages 41-44.
Unless specified, the models assumed vague (low information) prior distributions (Kéry and Schaub, 2011, p. 36). The posterior distributions were estimated from a minimum of 1,000 Markov Chain Monte Carlo (MCMC) samples thinned from the second halves of three chains (Kéry and Schaub, 2011, pp. 38-40). Model convergence was confirmed by ensuring that Rhat (Kéry and Schaub, 2011, p. 40) was less than 1.1 for each of the parameters in the model (Kéry and Schaub, 2011, p. 61). Model adequacy was confirmed by examination of residual plots.
The posterior distributions of the fixed (Kéry and Schaub, 2011, p. 75) parameters are summarised in terms of a point estimate (mean), lower and upper 95% credible limits (2.5th and 97.5th percentiles), the standard deviation (SD), percent relative error (half the 95% credible interval as a percent of the point estimate) and significance (Kéry and Schaub, 2011, p. 37,42).
The results are displayed graphically by plotting the modeled relationships between particular variables and the response with 95% credible intervals (CRIs) with the remaining variables held constant. In general, continuous and discrete fixed variables are held constant at their mean and first level values respectively while random variables are held constant at their typical values (expected values of the underlying hyperdistributions) (Kéry and Schaub, 2011, pp. 77-82). Where informative the influence of particular variables is expressed in terms of the effect size (i.e., percent change in the response variable) with 95% credible intervals (Bradford et al. 2005). Plots were produced using the ggplot2 R package (Wickham, 2009).
Observer Efficiency
The aerial observer efficiency was estimated from ground counts using a Poisson model. Key assumptions of the observer efficiency model include:
- The spawner abundance at a site does not change between the ground and aerial count.
- The ground observers are unbiased with the ground count being drawn from a Poisson distribution.
- The aerial observer efficiency does not vary systematically with abundance or channel type, i.e., sidechannel versus main channel.
- The aerial counts are drawn from an overdispersed Poisson distribution.
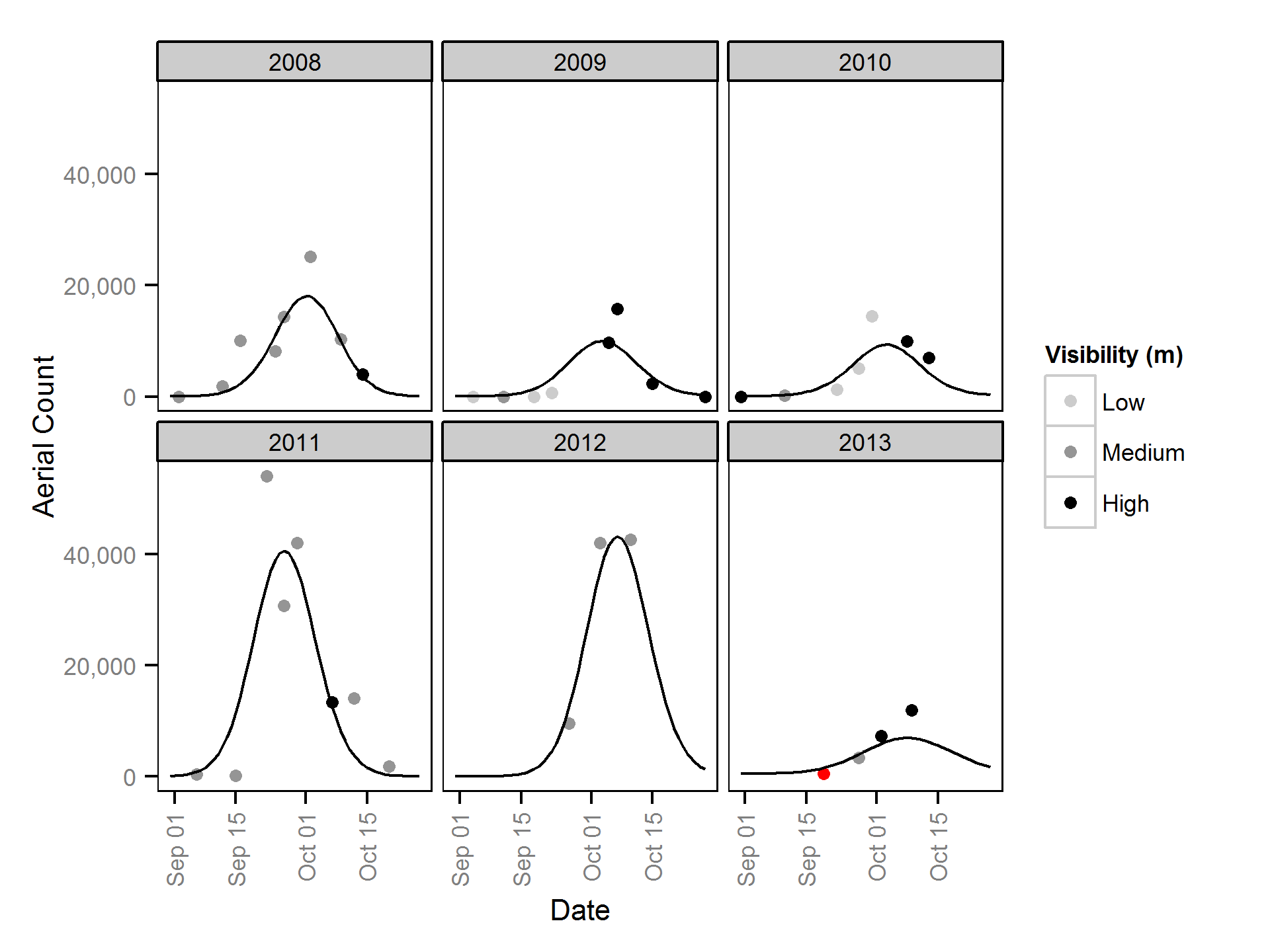
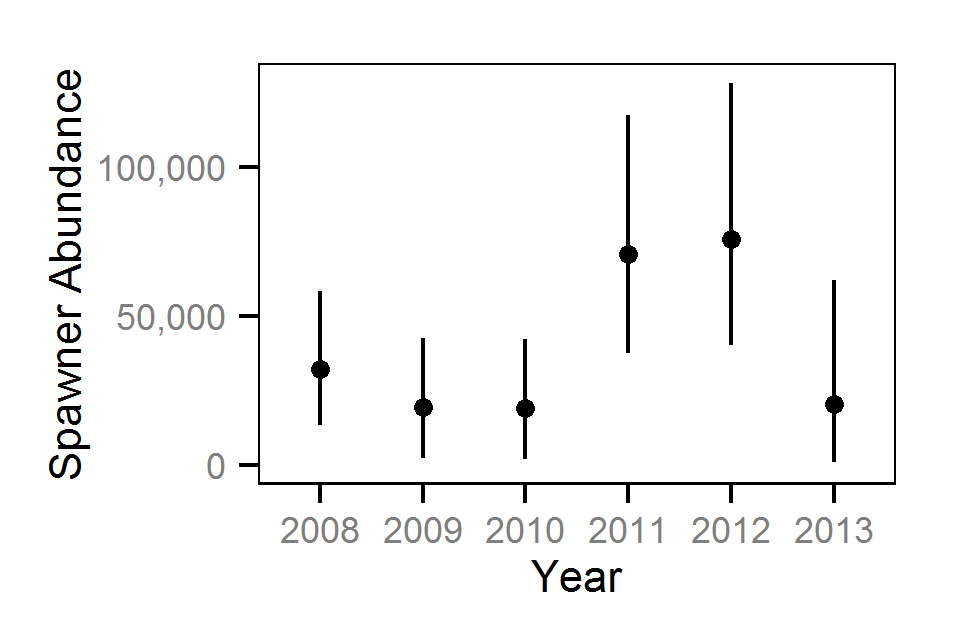
Area-Under-the-Curve
The aerial spawner counts were analysed using a hierarchical Bayesian Area-Under-the-Curve (AUC) model (Hilborn et al. 1999). Key assumptions of the AUC model include:
- Spawner arrival and departure are normally distributed (Hilborn et al. 1999)
- The duration of spawning is constant across years
- The peak spawn timing in each year is drawn from a normal distribution (Su et al. 2001)
- The residence time is between 7 and 14 days for spawners (Acara, 1970; Morbey and Ydenberg, 2003)
- The observer efficiency is described by a normal distribution with a mean of 1.07 and SD of 0.19 and lower and upper limits of 0.75 and 1.5 as estimated by the observer efficiency model
- The residual variation in the spawner counts is normally distributed
Model Code
The first three tables describe the JAGS distributions, functions and operators used in the models. For additional information on the JAGS dialect of the BUGS language see the JAGS User Manual (Plummer, 2012).
JAGS Distributions
| Distribution | Description |
|---|---|
dgamma(shape, rate) |
Gamma distribution |
dnorm(mu, sd^-2) |
Normal distribution |
dpois(lambda) |
Poisson distribution |
dunif(a, b) |
Uniform distribution |
JAGS Functions
| Function | Description |
|---|---|
length(x) |
Length of vector x |
log(x) |
Natural logarithm of x |
phi(x) |
Standard normal cumulative distribution function for x |
T(x,y) |
Truncate distribution so that values lie between x and y |
JAGS Operators
| Operator | Description |
|---|---|
<- |
Deterministic relationship |
~ |
Stochastic relationship |
1:n |
Vector of integers from 1 to n |
a[1:n] |
Subset of first n values in a |
for (i in 1:n) {...} |
Repeat … for 1 to n times incrementing i each time |
x^y |
Power where x is raised to the power of y |
Observer Efficiency
| Variable/Parameter | Description |
|---|---|
Aerial[i] |
Aerial count for ith survey |
bAbundance |
Intercept for log(eAbundance) |
bDispersion |
Variation in eDispersion |
bEfficiency |
Intercept for eEfficiency |
eAbundance[i] |
Predicted spawner abundance for ith survey |
eAerial[i] |
Predicted aerial count for ith survey |
eEfficiency[i] |
Predicted aerial observer efficiency for ith survey |
Ground[i] |
Ground count for ith survey |
sAbundance |
SD of log(eAbundance) |
Observer Efficiency - Model 1
model {
bEfficiency ~ dunif(0, 3)
bAbundance ~ dnorm(5, 5^-2)
sAbundance ~ dunif(0, 5)
bDispersion ~ dgamma(0.1, 0.1)
for (i in 1:length(Aerial)) {
eEfficiency[i] <- bEfficiency
eAbundance[i] ~ dlnorm(bAbundance, sAbundance^-2)
Ground[i] ~ dpois(eAbundance[i])
eAerial[i] <- eAbundance[i] * eEfficiency[i]
eDispersion[i] ~ dgamma(bDispersion, bDispersion)
Aerial[i] ~ dpois(eAerial[i] * eDispersion[i])
}
}Area-Under-The-Curve
| Variable/Parameter | Description |
|---|---|
bAbundance |
Intercept for log(eAbundance) |
bAbundanceYear[i] |
Effect of ith year on log(eAbundance) |
bDuration |
Intercept for eDuration |
bEfficiency |
Intercept for eEfficiency |
bPeakTiming |
Intercept for ePeakTiming |
bPeakTimingYear[i] |
Effect of ith year on ePeakTiming |
bResidenceTime |
Spawner residence time |
Count[i] |
Spawner count on ith survey |
Dayte[i] |
Centred day of the year of ith survey |
eAbundance[i] |
Predicted annual spawner abundance for ith survey |
eCount[i] |
Predicted spawner count on ith survey |
eDuration[i] |
Expected SD of the duration of spawner arrival timing |
eEfficiency[i] |
Predicted aerial observer efficiency ith survey |
ePeakTiming[i] |
Predicted timing of annual peak spawner abundance for ith survey |
eSpawners[i] |
Predicted number of spawners on ith survey |
sCount |
SD of log(eSpawners) |
Area-Under-The-Curve - Model 1
model {
bEfficiency ~ dnorm(1.07, 0.19^-2) T(0.75, 1.5)
bAbundance ~ dnorm(10, 5^-2)
bPeakTiming ~ dnorm(0, 5)
bDuration ~ dunif(0, 42)
sAbundanceYear ~ dunif(0, 5)
sPeakTimingYear ~ dunif(0, 28)
for (i in 1:nYear) {
bAbundanceYear[i] ~ dnorm (0, sAbundanceYear^-2)
bPeakTimingYear[i] ~ dnorm (0, sPeakTimingYear^-2)
}
bResidenceTime ~ dunif(7, 14)
sCount ~ dunif(0, 10000)
for (i in 1:length(Count)) {
log(eAbundance[i]) <- bAbundance
+ bAbundanceYear[Year[i]]
ePeakTiming[i] <- bPeakTiming
+ bPeakTimingYear[Year[i]]
eDuration[i] <- bDuration
eSpawners[i] <- (phi((Dayte[i] - (ePeakTiming[i] - bResidenceTime/2))
/ eDuration[i])
- phi((Dayte[i] - (ePeakTiming[i] + bResidenceTime/2))
/ eDuration[i]))
* eAbundance[i]
eEfficiency[i] <- bEfficiency
eCount[i] <- eSpawners[i] * eEfficiency[i]
Count[i] ~ dnorm(eCount[i], sCount^-2)
}
} Parameter Estimates
The posterior distributions for the fixed (Kery and Schaub 2011 p. 75) parameters in each model are summarised below.
Observer Efficiency
| Parameter | Estimate | Lower | Upper | SD | Error | Significance |
|---|---|---|---|---|---|---|
| bAbundance | 5.094 | 3.914 | 6.301 | 0.6076 | 23 | 0 |
| bDispersion | 3.617 | 1.129 | 7.692 | 1.7146 | 91 | 0 |
| bEfficiency | 1.071 | 0.751 | 1.500 | 0.1901 | 35 | 0 |
| sAbundance | 1.890 | 1.153 | 3.089 | 0.5120 | 51 | 0 |
| Rhat | Iterations |
|---|---|
| 1.1 | 40000 |
Area-Under-The-Curve
| Parameter | Estimate | Lower | Upper | SD | Error | Significance |
|---|---|---|---|---|---|---|
| bAbundance | 1.025e+01 | 8.5323 | 11.468 | 0.6888 | 14 | 0.0000 |
| bDuration | 6.155e+00 | 3.0483 | 8.540 | 1.3507 | 45 | 0.0000 |
| bEfficiency | 1.071e+00 | 0.7880 | 1.395 | 0.1632 | 28 | 0.0000 |
| bPeakTiming | 9.212e-02 | -0.7447 | 1.006 | 0.4385 | 950 | 0.8703 |
| bResidenceTime | 1.068e+01 | 7.2022 | 13.847 | 2.0605 | 31 | 0.0000 |
| sAbundanceYear | 1.166e+00 | 0.4239 | 3.204 | 0.6994 | 119 | 0.0000 |
| sCount | 6.495e+03 | 4819.4913 | 8677.541 | 992.4000 | 30 | 0.0000 |
| sPeakTimingYear | 1.013e+01 | 4.3513 | 22.884 | 4.7219 | 92 | 0.0000 |
| Rhat | Iterations |
|---|---|
| 1.03 | 1e+05 |
Figures
Observer Efficiency
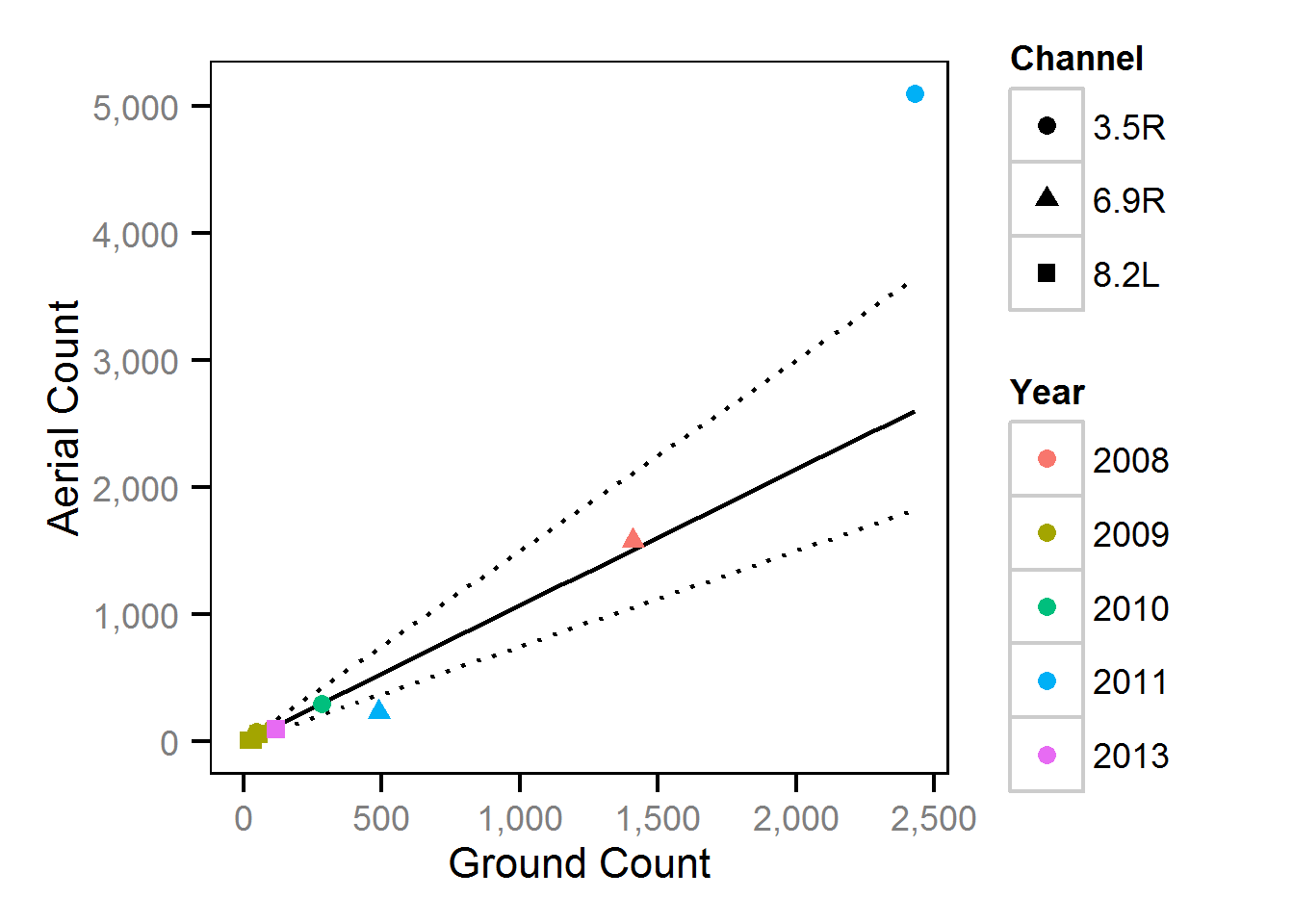
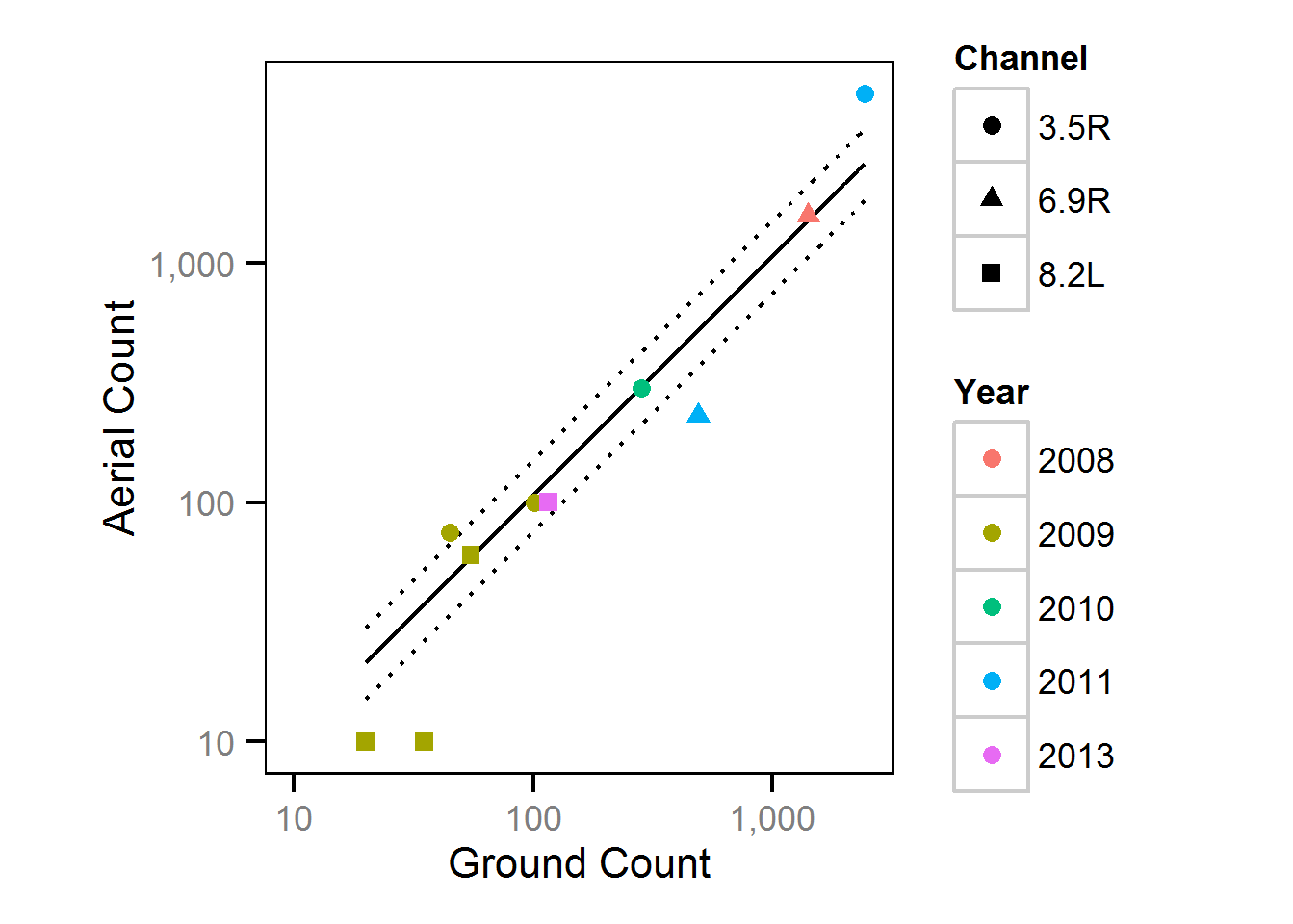
Area-Under-The-Curve
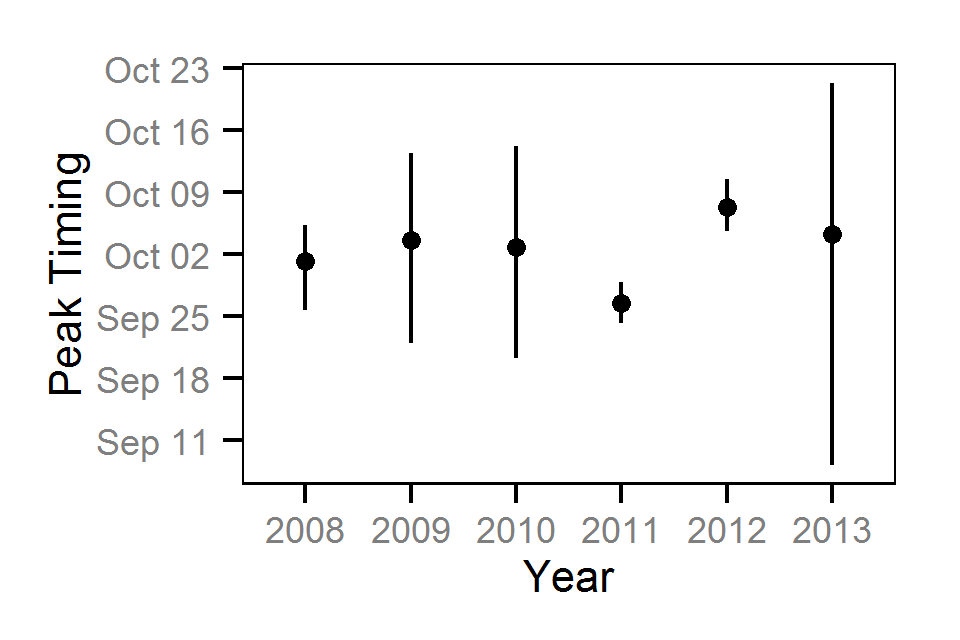
References
- {A.H.} Acara, (1970) The Meadow Creek Spawning Channel. Unpublished Report..
- Michael Bradford, Josh Korman, Paul Higgins, (2005) Using confidence intervals to estimate the response of salmon populations (Oncorhynchus spp.) to experimental habitat alterations. Canadian Journal of Fisheries and Aquatic Sciences 62 (12) 2716-2726 10.1139/f05-179
- Hadley Wickham, (2009) ggplot2: elegant graphics for data analysis. http://had.co.nz/ggplot2/book
- Ray Hilborn, Brian Bue, Samuel Sharr, (1999) Estimating spawning escapements from periodic counts: a comparison of methods. Canadian Journal of Fisheries and Aquatic Sciences 56 (5) 888-896 10.1139/f99-013
- Joseph Thorley, (2014) jaggernaut: An R package to facilitate Bayesian analyses using JAGS (Just Another Gibbs Sampler). https://github.com/joethorley/jaggernaut
- Marc Kéry, Michael Schaub, (2011) Bayesian population analysis using {WinBUGS} : a hierarchical perspective. http://www.vogelwarte.ch/bpa.html
- Y. Morbey, R. Ydenberg, (2003) Timing games in the reproductive phenology of female Pacific salmon (Oncorhynchus spp.). The American Naturalist 161 (2) 284–298-NA http://www.jstor.org/stable/10.1086/345785
- Martyn Plummer, (2012) {JAGS} version 3.3.0 user manual. http://sourceforge.net/projects/mcmc-jags/files/Manuals/3.x/
- R Team, (2013) R: a language and environment for statistical computing. http://www.R-project.org
- Zhenming Su, Milo Adkison, Benjamin Alen, (2001) A hierarchical Bayesian model for estimating historical salmon escapement and escapement timing. Canadian Journal of Fisheries and Aquatic Sciences 58 (8) 1648-1662 10.1139/cjfas-58-8-1648
Acknowledgements
This analysis report was made possible through the contributions of the following organisations and individuals:
- BC Hydro
- Guy Martel
- Okanagan National Alliance
- Michael Zimmer
- Natasha Audy
- Skyeler Folks
- LGL Limited
- Elmar Plate
- Amec Earth and Environmental
- Louise Porto
- Crystal Lawrence
- Clint Tarala
- Poisson Consulting Ltd.
- Robyn Irvine